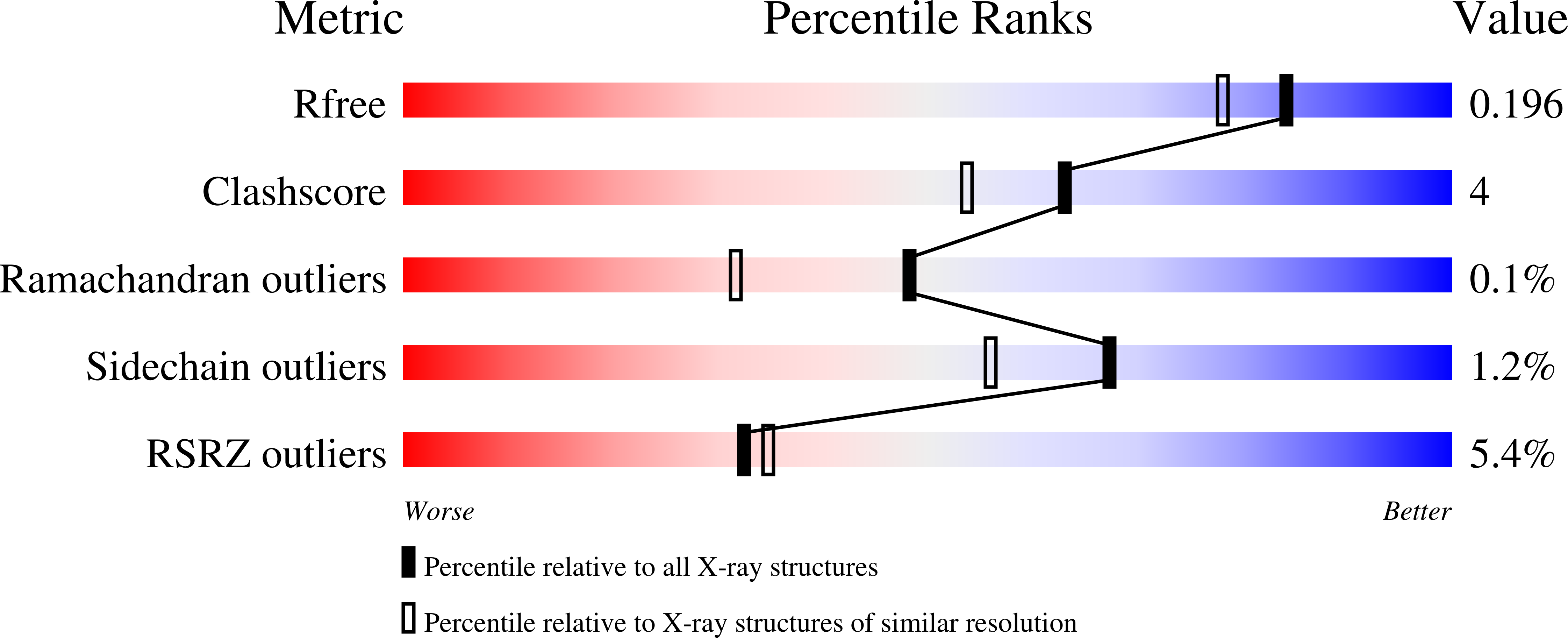
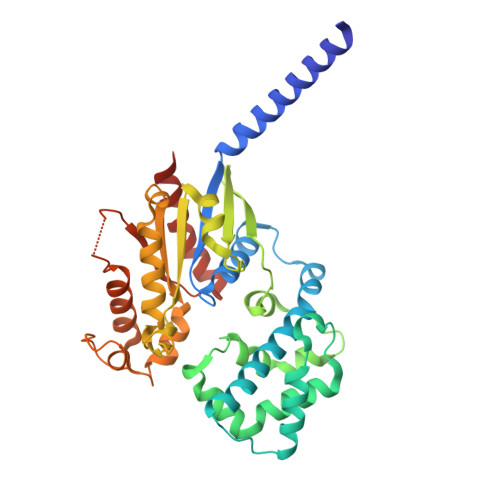
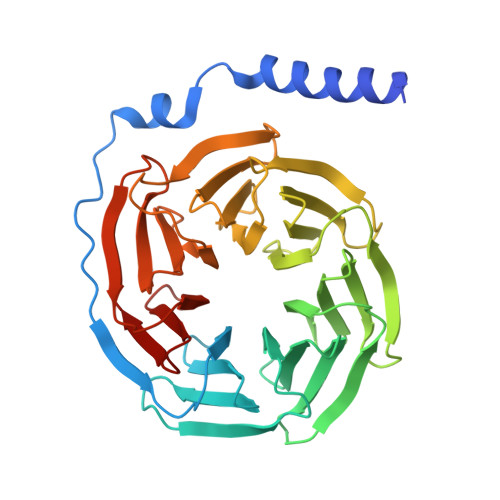
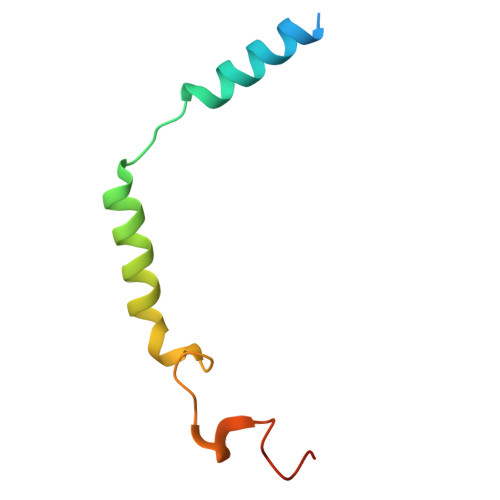
Cyclic peptide inhibitors function as molecular glues to stabilize Gq/11 heterotrimers.
Muhle, J., Alenfelder, J., Rodrigues, M.J., Jurgenliemke, L., Guixa-Gonzalez, R., Gratz, L., Andres, F., Bacchin, A., Hennig, M., Schihada, H., Crusemann, M., Konig, G.M., Schertler, G., Kostenis, E., Deupi, X.(2025) Proc Natl Acad Sci U S A 122: e2418398122-e2418398122
- PubMed: 40333756
- DOI: https://doi.org/10.1073/pnas.2418398122
- Primary Citation of Related Structures:
8QEG, 8QEH - PubMed Abstract:
Heterotrimeric Gα:Gβγ G proteins function as molecular switches downstream of G protein-coupled receptors (GPCRs). They alternate between a heterotrimeric GDP-bound OFF-state and a GTP-bound ON-state in which Gα GTP is separated from the Gβγ dimer. Consequently, pharmacological tools to securely prevent the OFF-ON transition are of utmost importance to investigate their molecular switch function, specific contribution to GPCR signal transduction, and potential as drug targets. FR900359 (FR) and YM-254890 (YM), two natural cyclic peptides and highly specific inhibitors of Gq/11 heterotrimers, are exactly such tools. To date, their efficient and long-lasting inhibition of Gq/11 signaling has been attributed solely to a wedge-like binding to Gα, thereby preventing separation of the GTPase and α-helical domains and thus GDP release. Here, we use X-ray crystallography, biochemical and signaling assays, and BRET-based biosensors to show that FR and YM also function as stabilizers of the Gα:Gβγ subunit interface. Our high-resolution structures reveal a network of residues in Gα and two highly conserved amino acids in Gβ that are targeted by FR and YM to glue the Gβγ complex to the inactive Gα GDP subunit. Unlike all previously developed nucleotide-state specific inhibitors that sequester Gα in its OFF-state but compete with Gβγ, FR and YM actively promote the inhibitory occlusion of Gα GDP by Gβγ. In doing so, they securely lock the entire heterotrimer, not just Gα, in its inactive state. Our results identify FR and YM as molecular glues for Gα and Gβγ that combine simultaneous binding to both subunits with inhibition of G protein signaling.
Organizational Affiliation:
Laboratory of Biomolecular Research, PSI Center for Life Sciences, Villigen 5232, Switzerland.