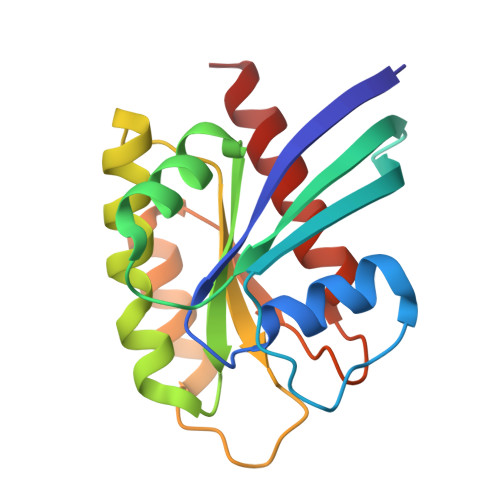
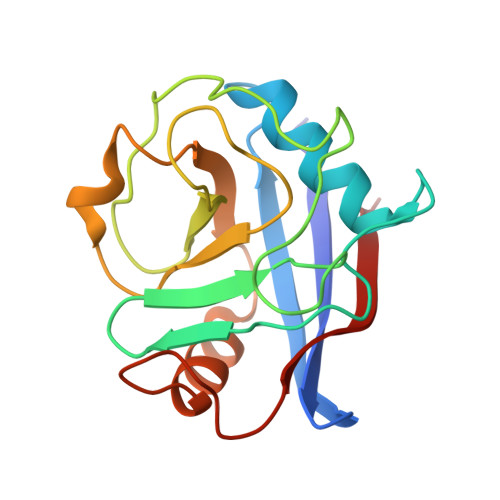
A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Weller, C., Burnett, G.L., Jiang, L., Chakraborty, S., Zhang, D., Vita, N.A., Dilly, J., Kim, E., Maldonato, B., Seamon, K., Eilerts, D.F., Milin, A., Marquez, A., Spradlin, J., Helland, C., Gould, A., Ziv, T.B., Dinh, P., Steele, S.L., Wang, Z., Mu, Y., Chugh, S., Feng, H., Hennessey, C., Wang, J., Roth, J., Rees, M., Ronan, M., Wolpin, B.M., Hahn, W.C., Holderfield, M., Wang, Z., Koltun, E.S., Singh, M., Gill, A.L., Smith, J.A.M., Aguirre, A.J., Jiang, J., Knox, J.E., Wildes, D.(2025) Science 389: eads0239-eads0239
- PubMed: 40705880
- DOI: https://doi.org/10.1126/science.ads0239
- Primary Citation of Related Structures:
9CT7, 9CT8, 9CT9, 9CTA, 9CTB, 9E3S - PubMed Abstract:
Mutant RAS proteins are among the most prevalent drivers of human cancer, and the glycine to aspartic acid mutation at codon 12 (G12D) is the most common variant. Mutation-selective covalent inhibitors spare RAS in healthy tissue and enable extended pharmacodynamic effect, but covalent targeting of RAS G12D is hindered by low nucleophilicity and high proteomic abundance of carboxylic acids. We overcame these challenges with compounds that bind cyclophilin A (CYPA) to create a neomorphic protein-protein interface between CYPA and active RAS that enables selective, enzyme-like rate enhancement of the covalent reaction between D12 and electrophilic warheads with exceptionally low intrinsic reactivity. This approach yielded orally bioavailable compounds with marked antitumor activity in multiple preclinical models of KRAS G12D cancers, including the investigational agent zoldonrasib (RMC-9805) currently undergoing clinical evaluation (NCT06040541).
Organizational Affiliation:
Revolution Medicines, Inc., Redwood City, CA, USA.